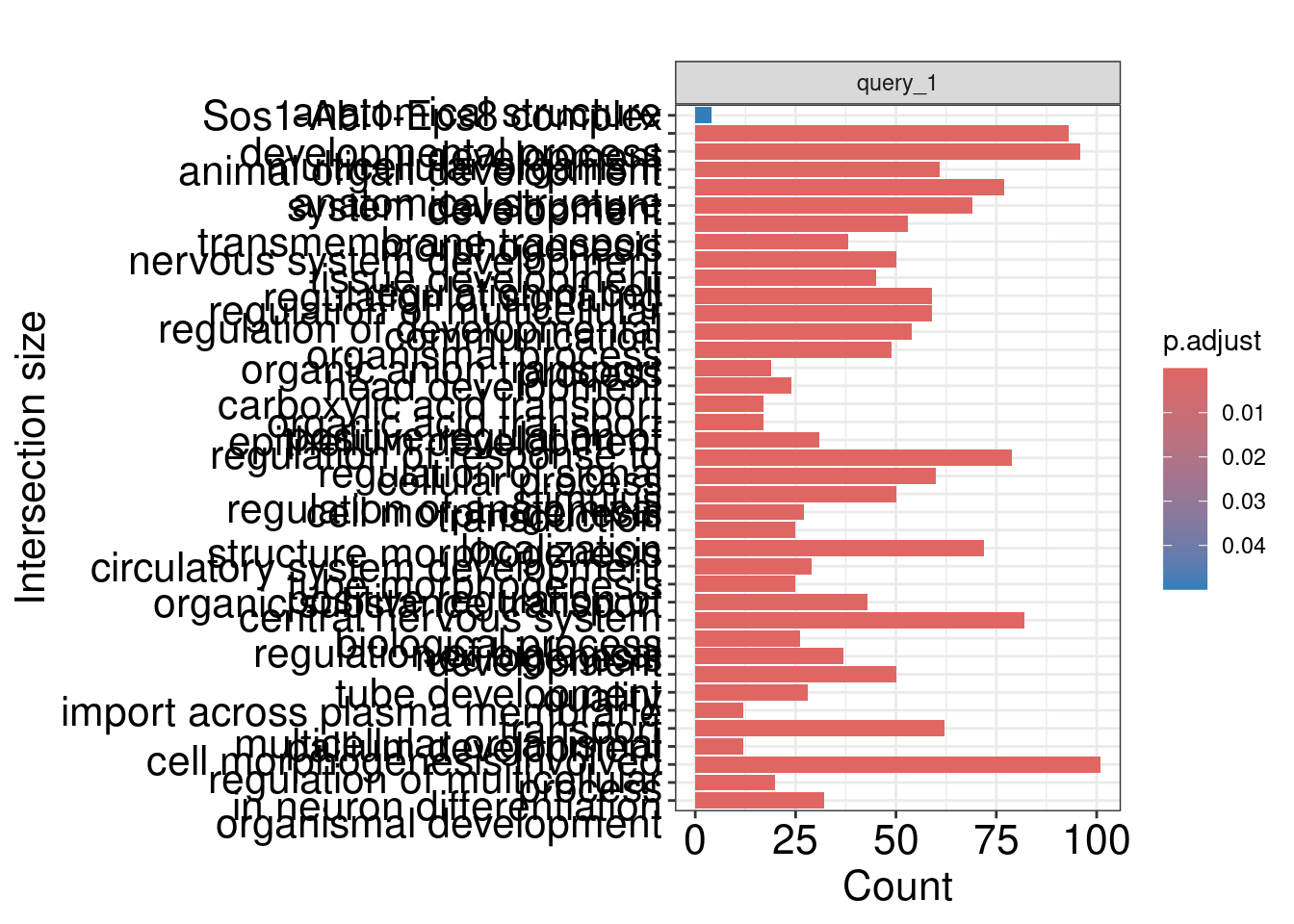
| CORUM:141 |
query_1 |
CORUM |
CORUM:141 |
Sos1-Abi1-Eps8 complex |
0.0499301 |
14 |
2 |
3 |
1082 |
2/14 |
3/1082 |
| CORUM:3143 |
query_1 |
CORUM |
CORUM:3143 |
Sos1-Abi1-Eps8 complex |
0.0499301 |
14 |
2 |
3 |
1082 |
2/14 |
3/1082 |
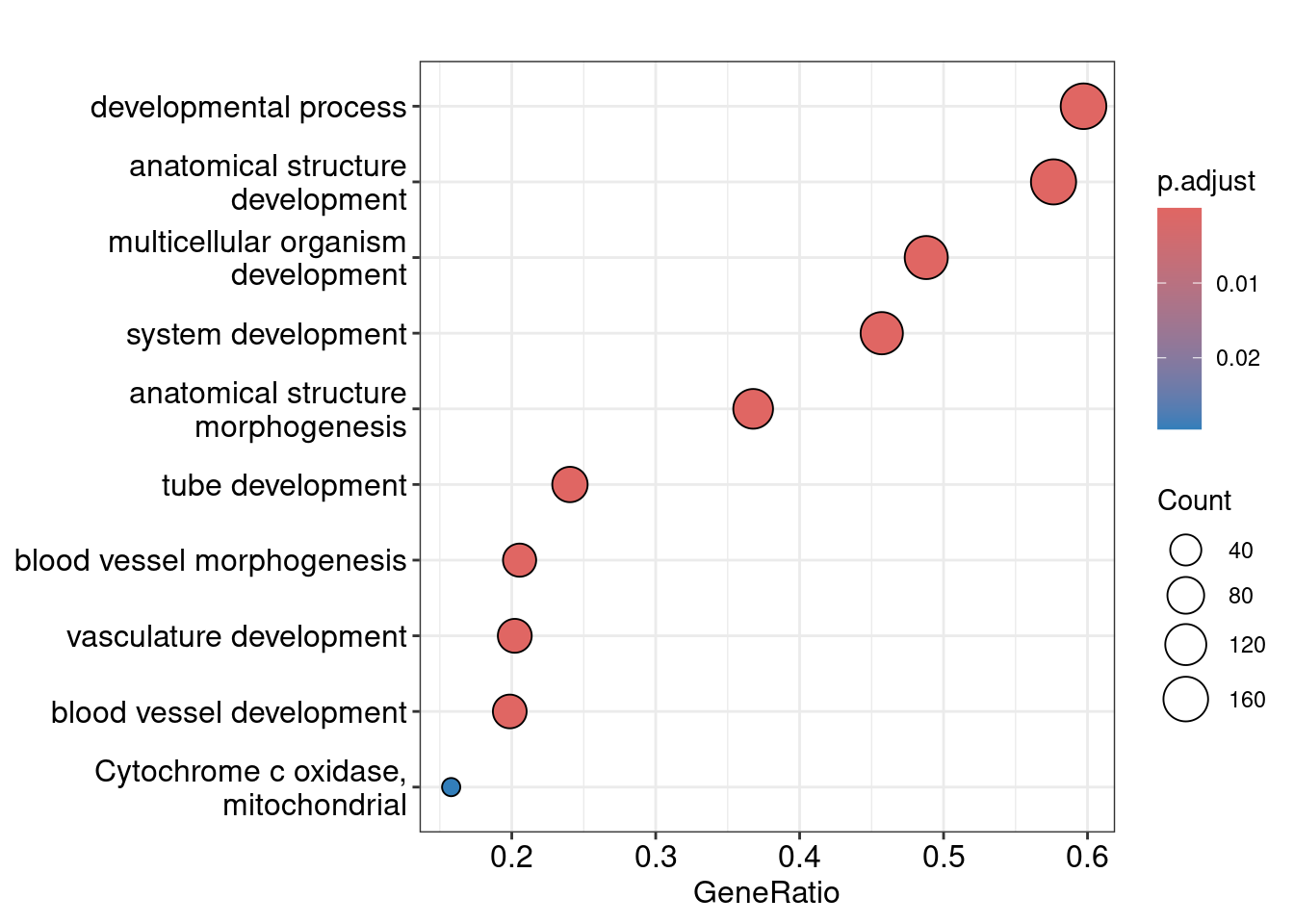
| GO:0048856 |
query_1 |
GO:BP |
GO:0048856 |
anatomical structure development |
0.0000000 |
190 |
93 |
6269 |
26944 |
93/190 |
6269/26944 |
| GO:0032502 |
query_1 |
GO:BP |
GO:0032502 |
developmental process |
0.0000000 |
190 |
96 |
6854 |
26944 |
96/190 |
6854/26944 |
| GO:0048513 |
query_1 |
GO:BP |
GO:0048513 |
animal organ development |
0.0000000 |
190 |
61 |
3260 |
26944 |
61/190 |
3260/26944 |
| GO:0007275 |
query_1 |
GO:BP |
GO:0007275 |
multicellular organism development |
0.0000000 |
190 |
77 |
4879 |
26944 |
77/190 |
4879/26944 |
| GO:0048731 |
query_1 |
GO:BP |
GO:0048731 |
system development |
0.0000000 |
190 |
69 |
4117 |
26944 |
69/190 |
4117/26944 |
| GO:0009653 |
query_1 |
GO:BP |
GO:0009653 |
anatomical structure morphogenesis |
0.0000000 |
190 |
53 |
2761 |
26944 |
53/190 |
2761/26944 |
| GO:0055085 |
query_1 |
GO:BP |
GO:0055085 |
transmembrane transport |
0.0000000 |
190 |
38 |
1525 |
26944 |
38/190 |
1525/26944 |
| GO:0007399 |
query_1 |
GO:BP |
GO:0007399 |
nervous system development |
0.0000000 |
190 |
50 |
2529 |
26944 |
50/190 |
2529/26944 |
| GO:0009888 |
query_1 |
GO:BP |
GO:0009888 |
tissue development |
0.0000000 |
190 |
45 |
2122 |
26944 |
45/190 |
2122/26944 |
| GO:0023051 |
query_1 |
GO:BP |
GO:0023051 |
regulation of signaling |
0.0000004 |
190 |
59 |
3616 |
26944 |
59/190 |
3616/26944 |
| GO:0010646 |
query_1 |
GO:BP |
GO:0010646 |
regulation of cell communication |
0.0000004 |
190 |
59 |
3616 |
26944 |
59/190 |
3616/26944 |
| GO:0051239 |
query_1 |
GO:BP |
GO:0051239 |
regulation of multicellular organismal process |
0.0000006 |
190 |
54 |
3165 |
26944 |
54/190 |
3165/26944 |
| GO:0050793 |
query_1 |
GO:BP |
GO:0050793 |
regulation of developmental process |
0.0000006 |
190 |
49 |
2698 |
26944 |
49/190 |
2698/26944 |
| GO:0015711 |
query_1 |
GO:BP |
GO:0015711 |
organic anion transport |
0.0000016 |
190 |
19 |
464 |
26944 |
19/190 |
464/26944 |
| GO:0060322 |
query_1 |
GO:BP |
GO:0060322 |
head development |
0.0000026 |
190 |
24 |
778 |
26944 |
24/190 |
778/26944 |
| GO:0046942 |
query_1 |
GO:BP |
GO:0046942 |
carboxylic acid transport |
0.0000039 |
190 |
17 |
384 |
26944 |
17/190 |
384/26944 |
| GO:0015849 |
query_1 |
GO:BP |
GO:0015849 |
organic acid transport |
0.0000042 |
190 |
17 |
386 |
26944 |
17/190 |
386/26944 |
| GO:0060429 |
query_1 |
GO:BP |
GO:0060429 |
epithelium development |
0.0000053 |
190 |
31 |
1309 |
26944 |
31/190 |
1309/26944 |
| GO:0048522 |
query_1 |
GO:BP |
GO:0048522 |
positive regulation of cellular process |
0.0000059 |
190 |
79 |
6078 |
26944 |
79/190 |
6078/26944 |
| GO:0048583 |
query_1 |
GO:BP |
GO:0048583 |
regulation of response to stimulus |
0.0000062 |
190 |
60 |
3987 |
26944 |
60/190 |
3987/26944 |
| GO:0009966 |
query_1 |
GO:BP |
GO:0009966 |
regulation of signal transduction |
0.0000080 |
190 |
50 |
3005 |
26944 |
50/190 |
3005/26944 |
| GO:0000902 |
query_1 |
GO:BP |
GO:0000902 |
cell morphogenesis |
0.0000102 |
190 |
27 |
1045 |
26944 |
27/190 |
1045/26944 |
| GO:0022603 |
query_1 |
GO:BP |
GO:0022603 |
regulation of anatomical structure morphogenesis |
0.0000104 |
190 |
25 |
904 |
26944 |
25/190 |
904/26944 |
| GO:0051179 |
query_1 |
GO:BP |
GO:0051179 |
localization |
0.0000120 |
190 |
72 |
5364 |
26944 |
72/190 |
5364/26944 |
| GO:0072359 |
query_1 |
GO:BP |
GO:0072359 |
circulatory system development |
0.0000157 |
190 |
29 |
1216 |
26944 |
29/190 |
1216/26944 |
| GO:0035239 |
query_1 |
GO:BP |
GO:0035239 |
tube morphogenesis |
0.0000171 |
190 |
25 |
927 |
26944 |
25/190 |
927/26944 |
| GO:0071702 |
query_1 |
GO:BP |
GO:0071702 |
organic substance transport |
0.0000195 |
190 |
43 |
2419 |
26944 |
43/190 |
2419/26944 |
| GO:0048518 |
query_1 |
GO:BP |
GO:0048518 |
positive regulation of biological process |
0.0000196 |
190 |
82 |
6589 |
26944 |
82/190 |
6589/26944 |
| GO:0007417 |
query_1 |
GO:BP |
GO:0007417 |
central nervous system development |
0.0000201 |
190 |
26 |
1006 |
26944 |
26/190 |
1006/26944 |
| GO:0022008 |
query_1 |
GO:BP |
GO:0022008 |
neurogenesis |
0.0000275 |
190 |
37 |
1907 |
26944 |
37/190 |
1907/26944 |
| GO:0065008 |
query_1 |
GO:BP |
GO:0065008 |
regulation of biological quality |
0.0000319 |
190 |
50 |
3134 |
26944 |
50/190 |
3134/26944 |
| GO:0035295 |
query_1 |
GO:BP |
GO:0035295 |
tube development |
0.0000330 |
190 |
28 |
1181 |
26944 |
28/190 |
1181/26944 |
| GO:0098739 |
query_1 |
GO:BP |
GO:0098739 |
import across plasma membrane |
0.0000375 |
190 |
12 |
201 |
26944 |
12/190 |
201/26944 |
| GO:0006810 |
query_1 |
GO:BP |
GO:0006810 |
transport |
0.0000380 |
190 |
62 |
4397 |
26944 |
62/190 |
4397/26944 |
| GO:0021543 |
query_1 |
GO:BP |
GO:0021543 |
pallium development |
0.0000548 |
190 |
12 |
208 |
26944 |
12/190 |
208/26944 |
| GO:0032501 |
query_1 |
GO:BP |
GO:0032501 |
multicellular organismal process |
0.0000598 |
190 |
101 |
9132 |
26944 |
101/190 |
9132/26944 |
| GO:0048667 |
query_1 |
GO:BP |
GO:0048667 |
cell morphogenesis involved in neuron differentiation |
0.0000640 |
190 |
20 |
647 |
26944 |
20/190 |
647/26944 |
| GO:2000026 |
query_1 |
GO:BP |
GO:2000026 |
regulation of multicellular organismal development |
0.0000737 |
190 |
32 |
1550 |
26944 |
32/190 |
1550/26944 |
| GO:0048858 |
query_1 |
GO:BP |
GO:0048858 |
cell projection morphogenesis |
0.0000786 |
190 |
21 |
721 |
26944 |
21/190 |
721/26944 |
| GO:0008283 |
query_1 |
GO:BP |
GO:0008283 |
cell population proliferation |
0.0000803 |
190 |
39 |
2168 |
26944 |
39/190 |
2168/26944 |
| GO:0007420 |
query_1 |
GO:BP |
GO:0007420 |
brain development |
0.0000824 |
190 |
21 |
723 |
26944 |
21/190 |
723/26944 |
| GO:0048870 |
query_1 |
GO:BP |
GO:0048870 |
cell motility |
0.0001296 |
190 |
34 |
1760 |
26944 |
34/190 |
1760/26944 |
| GO:0034330 |
query_1 |
GO:BP |
GO:0034330 |
cell junction organization |
0.0001363 |
190 |
22 |
814 |
26944 |
22/190 |
814/26944 |
| GO:0032989 |
query_1 |
GO:BP |
GO:0032989 |
cellular anatomical entity morphogenesis |
0.0001684 |
190 |
22 |
824 |
26944 |
22/190 |
824/26944 |
| GO:0051234 |
query_1 |
GO:BP |
GO:0051234 |
establishment of localization |
0.0001837 |
190 |
63 |
4696 |
26944 |
63/190 |
4696/26944 |
| GO:0016477 |
query_1 |
GO:BP |
GO:0016477 |
cell migration |
0.0002107 |
190 |
31 |
1538 |
26944 |
31/190 |
1538/26944 |
| GO:0042127 |
query_1 |
GO:BP |
GO:0042127 |
regulation of cell population proliferation |
0.0002180 |
190 |
34 |
1799 |
26944 |
34/190 |
1799/26944 |
| GO:0048812 |
query_1 |
GO:BP |
GO:0048812 |
neuron projection morphogenesis |
0.0002235 |
190 |
20 |
699 |
26944 |
20/190 |
699/26944 |
| GO:0120039 |
query_1 |
GO:BP |
GO:0120039 |
plasma membrane bounded cell projection morphogenesis |
0.0003136 |
190 |
20 |
714 |
26944 |
20/190 |
714/26944 |
| GO:0006869 |
query_1 |
GO:BP |
GO:0006869 |
lipid transport |
0.0003207 |
190 |
16 |
459 |
26944 |
16/190 |
459/26944 |
| GO:0006811 |
query_1 |
GO:BP |
GO:0006811 |
monoatomic ion transport |
0.0003327 |
190 |
27 |
1239 |
26944 |
27/190 |
1239/26944 |
| GO:0042592 |
query_1 |
GO:BP |
GO:0042592 |
homeostatic process |
0.0003471 |
190 |
34 |
1835 |
26944 |
34/190 |
1835/26944 |
| GO:1905039 |
query_1 |
GO:BP |
GO:1905039 |
carboxylic acid transmembrane transport |
0.0003553 |
190 |
10 |
159 |
26944 |
10/190 |
159/26944 |
| GO:1903825 |
query_1 |
GO:BP |
GO:1903825 |
organic acid transmembrane transport |
0.0003767 |
190 |
10 |
160 |
26944 |
10/190 |
160/26944 |
| GO:0021537 |
query_1 |
GO:BP |
GO:0021537 |
telencephalon development |
0.0005154 |
190 |
13 |
306 |
26944 |
13/190 |
306/26944 |
| GO:0050808 |
query_1 |
GO:BP |
GO:0050808 |
synapse organization |
0.0005369 |
190 |
17 |
539 |
26944 |
17/190 |
539/26944 |
| GO:0009967 |
query_1 |
GO:BP |
GO:0009967 |
positive regulation of signal transduction |
0.0005963 |
190 |
31 |
1613 |
26944 |
31/190 |
1613/26944 |
| GO:0031175 |
query_1 |
GO:BP |
GO:0031175 |
neuron projection development |
0.0007619 |
190 |
25 |
1131 |
26944 |
25/190 |
1131/26944 |
| GO:0010647 |
query_1 |
GO:BP |
GO:0010647 |
positive regulation of cell communication |
0.0007666 |
190 |
35 |
1990 |
26944 |
35/190 |
1990/26944 |
| GO:0023056 |
query_1 |
GO:BP |
GO:0023056 |
positive regulation of signaling |
0.0007666 |
190 |
35 |
1990 |
26944 |
35/190 |
1990/26944 |
| GO:0000165 |
query_1 |
GO:BP |
GO:0000165 |
MAPK cascade |
0.0008051 |
190 |
20 |
758 |
26944 |
20/190 |
758/26944 |
| GO:0042908 |
query_1 |
GO:BP |
GO:0042908 |
xenobiotic transport |
0.0008319 |
190 |
6 |
42 |
26944 |
6/190 |
42/26944 |
| GO:0007166 |
query_1 |
GO:BP |
GO:0007166 |
cell surface receptor signaling pathway |
0.0008615 |
190 |
43 |
2766 |
26944 |
43/190 |
2766/26944 |
| GO:0007267 |
query_1 |
GO:BP |
GO:0007267 |
cell-cell signaling |
0.0009186 |
190 |
32 |
1734 |
26944 |
32/190 |
1734/26944 |
| GO:0048871 |
query_1 |
GO:BP |
GO:0048871 |
multicellular organismal-level homeostasis |
0.0009436 |
190 |
22 |
912 |
26944 |
22/190 |
912/26944 |
| GO:0060249 |
query_1 |
GO:BP |
GO:0060249 |
anatomical structure homeostasis |
0.0009622 |
190 |
12 |
271 |
26944 |
12/190 |
271/26944 |
| GO:0001894 |
query_1 |
GO:BP |
GO:0001894 |
tissue homeostasis |
0.0009622 |
190 |
12 |
271 |
26944 |
12/190 |
271/26944 |
| GO:0021766 |
query_1 |
GO:BP |
GO:0021766 |
hippocampus development |
0.0016338 |
190 |
8 |
107 |
26944 |
8/190 |
107/26944 |
| GO:0010876 |
query_1 |
GO:BP |
GO:0010876 |
lipid localization |
0.0016348 |
190 |
16 |
519 |
26944 |
16/190 |
519/26944 |
| GO:0030154 |
query_1 |
GO:BP |
GO:0030154 |
cell differentiation |
0.0018528 |
190 |
61 |
4777 |
26944 |
61/190 |
4777/26944 |
| GO:0048869 |
query_1 |
GO:BP |
GO:0048869 |
cellular developmental process |
0.0018662 |
190 |
61 |
4778 |
26944 |
61/190 |
4778/26944 |
| GO:0015801 |
query_1 |
GO:BP |
GO:0015801 |
aromatic amino acid transport |
0.0019562 |
190 |
4 |
12 |
26944 |
4/190 |
12/26944 |
| GO:0002009 |
query_1 |
GO:BP |
GO:0002009 |
morphogenesis of an epithelium |
0.0024855 |
190 |
16 |
536 |
26944 |
16/190 |
536/26944 |
| GO:0048468 |
query_1 |
GO:BP |
GO:0048468 |
cell development |
0.0028680 |
190 |
46 |
3202 |
26944 |
46/190 |
3202/26944 |
| GO:0048699 |
query_1 |
GO:BP |
GO:0048699 |
generation of neurons |
0.0030070 |
190 |
30 |
1651 |
26944 |
30/190 |
1651/26944 |
| GO:0048646 |
query_1 |
GO:BP |
GO:0048646 |
anatomical structure formation involved in morphogenesis |
0.0030499 |
190 |
25 |
1221 |
26944 |
25/190 |
1221/26944 |
| GO:0051094 |
query_1 |
GO:BP |
GO:0051094 |
positive regulation of developmental process |
0.0032213 |
190 |
28 |
1480 |
26944 |
28/190 |
1480/26944 |
| GO:0061564 |
query_1 |
GO:BP |
GO:0061564 |
axon development |
0.0033074 |
190 |
16 |
548 |
26944 |
16/190 |
548/26944 |
| GO:0001568 |
query_1 |
GO:BP |
GO:0001568 |
blood vessel development |
0.0035696 |
190 |
19 |
761 |
26944 |
19/190 |
761/26944 |
| GO:0089718 |
query_1 |
GO:BP |
GO:0089718 |
amino acid import across plasma membrane |
0.0038184 |
190 |
6 |
54 |
26944 |
6/190 |
54/26944 |
| GO:0048519 |
query_1 |
GO:BP |
GO:0048519 |
negative regulation of biological process |
0.0047935 |
190 |
70 |
5966 |
26944 |
70/190 |
5966/26944 |
| GO:0048589 |
query_1 |
GO:BP |
GO:0048589 |
developmental growth |
0.0055980 |
190 |
19 |
785 |
26944 |
19/190 |
785/26944 |
| GO:0030030 |
query_1 |
GO:BP |
GO:0030030 |
cell projection organization |
0.0058156 |
190 |
30 |
1706 |
26944 |
30/190 |
1706/26944 |
| GO:0089709 |
query_1 |
GO:BP |
GO:0089709 |
L-histidine transmembrane transport |
0.0058894 |
190 |
3 |
5 |
26944 |
3/190 |
5/26944 |
| GO:0006812 |
query_1 |
GO:BP |
GO:0006812 |
monoatomic cation transport |
0.0060700 |
190 |
22 |
1022 |
26944 |
22/190 |
1022/26944 |
| GO:0051240 |
query_1 |
GO:BP |
GO:0051240 |
positive regulation of multicellular organismal process |
0.0062556 |
190 |
31 |
1804 |
26944 |
31/190 |
1804/26944 |
| GO:0001944 |
query_1 |
GO:BP |
GO:0001944 |
vasculature development |
0.0064772 |
190 |
19 |
793 |
26944 |
19/190 |
793/26944 |
| GO:0030900 |
query_1 |
GO:BP |
GO:0030900 |
forebrain development |
0.0066686 |
190 |
14 |
447 |
26944 |
14/190 |
447/26944 |
| GO:0007423 |
query_1 |
GO:BP |
GO:0007423 |
sensory organ development |
0.0068559 |
190 |
17 |
650 |
26944 |
17/190 |
650/26944 |
| GO:0048729 |
query_1 |
GO:BP |
GO:0048729 |
tissue morphogenesis |
0.0069967 |
190 |
17 |
651 |
26944 |
17/190 |
651/26944 |
| GO:1902531 |
query_1 |
GO:BP |
GO:1902531 |
regulation of intracellular signal transduction |
0.0070027 |
190 |
30 |
1722 |
26944 |
30/190 |
1722/26944 |
| GO:0048666 |
query_1 |
GO:BP |
GO:0048666 |
neuron development |
0.0077082 |
190 |
25 |
1287 |
26944 |
25/190 |
1287/26944 |
| GO:0048514 |
query_1 |
GO:BP |
GO:0048514 |
blood vessel morphogenesis |
0.0078977 |
190 |
17 |
657 |
26944 |
17/190 |
657/26944 |
| GO:0007155 |
query_1 |
GO:BP |
GO:0007155 |
cell adhesion |
0.0081522 |
190 |
27 |
1465 |
26944 |
27/190 |
1465/26944 |
| GO:0030001 |
query_1 |
GO:BP |
GO:0030001 |
metal ion transport |
0.0083653 |
190 |
20 |
884 |
26944 |
20/190 |
884/26944 |
| GO:0021761 |
query_1 |
GO:BP |
GO:0021761 |
limbic system development |
0.0099147 |
190 |
8 |
136 |
26944 |
8/190 |
136/26944 |
| GO:0043408 |
query_1 |
GO:BP |
GO:0043408 |
regulation of MAPK cascade |
0.0102169 |
190 |
17 |
670 |
26944 |
17/190 |
670/26944 |
| GO:0120036 |
query_1 |
GO:BP |
GO:0120036 |
plasma membrane bounded cell projection organization |
0.0105280 |
190 |
29 |
1666 |
26944 |
29/190 |
1666/26944 |
| GO:0006915 |
query_1 |
GO:BP |
GO:0006915 |
apoptotic process |
0.0116163 |
190 |
33 |
2050 |
26944 |
33/190 |
2050/26944 |
| GO:0032328 |
query_1 |
GO:BP |
GO:0032328 |
alanine transport |
0.0116984 |
190 |
4 |
18 |
26944 |
4/190 |
18/26944 |
| GO:0070887 |
query_1 |
GO:BP |
GO:0070887 |
cellular response to chemical stimulus |
0.0118233 |
190 |
44 |
3162 |
26944 |
44/190 |
3162/26944 |
| GO:0048584 |
query_1 |
GO:BP |
GO:0048584 |
positive regulation of response to stimulus |
0.0136077 |
190 |
36 |
2359 |
26944 |
36/190 |
2359/26944 |
| GO:0008610 |
query_1 |
GO:BP |
GO:0008610 |
lipid biosynthetic process |
0.0139009 |
190 |
17 |
686 |
26944 |
17/190 |
686/26944 |
| GO:0150104 |
query_1 |
GO:BP |
GO:0150104 |
transport across blood-brain barrier |
0.0147363 |
190 |
4 |
19 |
26944 |
4/190 |
19/26944 |
| GO:0035633 |
query_1 |
GO:BP |
GO:0035633 |
maintenance of blood-brain barrier |
0.0147363 |
190 |
4 |
19 |
26944 |
4/190 |
19/26944 |
| GO:0060856 |
query_1 |
GO:BP |
GO:0060856 |
establishment of blood-brain barrier |
0.0147363 |
190 |
4 |
19 |
26944 |
4/190 |
19/26944 |
| GO:0007409 |
query_1 |
GO:BP |
GO:0007409 |
axonogenesis |
0.0153363 |
190 |
14 |
481 |
26944 |
14/190 |
481/26944 |
| GO:0141124 |
query_1 |
GO:BP |
GO:0141124 |
intracellular signaling cassette |
0.0156398 |
190 |
30 |
1794 |
26944 |
30/190 |
1794/26944 |
| GO:0065007 |
query_1 |
GO:BP |
GO:0065007 |
biological regulation |
0.0166254 |
190 |
125 |
13527 |
26944 |
125/190 |
13527/26944 |
| GO:0050803 |
query_1 |
GO:BP |
GO:0050803 |
regulation of synapse structure or activity |
0.0169054 |
190 |
11 |
299 |
26944 |
11/190 |
299/26944 |
| GO:0010232 |
query_1 |
GO:BP |
GO:0010232 |
vascular transport |
0.0183190 |
190 |
4 |
20 |
26944 |
4/190 |
20/26944 |
| GO:0060562 |
query_1 |
GO:BP |
GO:0060562 |
epithelial tube morphogenesis |
0.0195003 |
190 |
12 |
363 |
26944 |
12/190 |
363/26944 |
| GO:0048523 |
query_1 |
GO:BP |
GO:0048523 |
negative regulation of cellular process |
0.0195045 |
190 |
65 |
5599 |
26944 |
65/190 |
5599/26944 |
| GO:0015718 |
query_1 |
GO:BP |
GO:0015718 |
monocarboxylic acid transport |
0.0201524 |
190 |
9 |
197 |
26944 |
9/190 |
197/26944 |
| GO:0051960 |
query_1 |
GO:BP |
GO:0051960 |
regulation of nervous system development |
0.0203891 |
190 |
15 |
562 |
26944 |
15/190 |
562/26944 |
| GO:0099173 |
query_1 |
GO:BP |
GO:0099173 |
postsynapse organization |
0.0210129 |
190 |
10 |
250 |
26944 |
10/190 |
250/26944 |
| GO:0007154 |
query_1 |
GO:BP |
GO:0007154 |
cell communication |
0.0214224 |
190 |
81 |
7582 |
26944 |
81/190 |
7582/26944 |
| GO:0030111 |
query_1 |
GO:BP |
GO:0030111 |
regulation of Wnt signaling pathway |
0.0229832 |
190 |
11 |
309 |
26944 |
11/190 |
309/26944 |
| GO:0012501 |
query_1 |
GO:BP |
GO:0012501 |
programmed cell death |
0.0244176 |
190 |
33 |
2125 |
26944 |
33/190 |
2125/26944 |
| GO:0008219 |
query_1 |
GO:BP |
GO:0008219 |
cell death |
0.0244176 |
190 |
33 |
2125 |
26944 |
33/190 |
2125/26944 |
| GO:0098655 |
query_1 |
GO:BP |
GO:0098655 |
monoatomic cation transmembrane transport |
0.0261546 |
190 |
18 |
797 |
26944 |
18/190 |
797/26944 |
| GO:0051897 |
query_1 |
GO:BP |
GO:0051897 |
positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction |
0.0266247 |
190 |
9 |
204 |
26944 |
9/190 |
204/26944 |
| GO:0048598 |
query_1 |
GO:BP |
GO:0048598 |
embryonic morphogenesis |
0.0270955 |
190 |
16 |
648 |
26944 |
16/190 |
648/26944 |
| GO:0030182 |
query_1 |
GO:BP |
GO:0030182 |
neuron differentiation |
0.0280434 |
190 |
27 |
1569 |
26944 |
27/190 |
1569/26944 |
| GO:0007507 |
query_1 |
GO:BP |
GO:0007507 |
heart development |
0.0297592 |
190 |
16 |
653 |
26944 |
16/190 |
653/26944 |
| GO:0045595 |
query_1 |
GO:BP |
GO:0045595 |
regulation of cell differentiation |
0.0299455 |
190 |
29 |
1761 |
26944 |
29/190 |
1761/26944 |
| GO:0006793 |
query_1 |
GO:BP |
GO:0006793 |
phosphorus metabolic process |
0.0310990 |
190 |
36 |
2450 |
26944 |
36/190 |
2450/26944 |
| GO:0030178 |
query_1 |
GO:BP |
GO:0030178 |
negative regulation of Wnt signaling pathway |
0.0325425 |
190 |
8 |
160 |
26944 |
8/190 |
160/26944 |
| GO:0042886 |
query_1 |
GO:BP |
GO:0042886 |
amide transport |
0.0349152 |
190 |
12 |
385 |
26944 |
12/190 |
385/26944 |
| GO:0033036 |
query_1 |
GO:BP |
GO:0033036 |
macromolecule localization |
0.0363057 |
190 |
42 |
3092 |
26944 |
42/190 |
3092/26944 |
| GO:0050789 |
query_1 |
GO:BP |
GO:0050789 |
regulation of biological process |
0.0406072 |
190 |
121 |
13144 |
26944 |
121/190 |
13144/26944 |
| GO:0120035 |
query_1 |
GO:BP |
GO:0120035 |
regulation of plasma membrane bounded cell projection organization |
0.0442460 |
190 |
17 |
751 |
26944 |
17/190 |
751/26944 |
| GO:0043067 |
query_1 |
GO:BP |
GO:0043067 |
regulation of programmed cell death |
0.0456833 |
190 |
28 |
1707 |
26944 |
28/190 |
1707/26944 |
| GO:0098657 |
query_1 |
GO:BP |
GO:0098657 |
import into cell |
0.0485926 |
190 |
19 |
916 |
26944 |
19/190 |
916/26944 |
| GO:0071944 |
query_1 |
GO:CC |
GO:0071944 |
cell periphery |
0.0000000 |
194 |
100 |
6862 |
26995 |
100/194 |
6862/26995 |
| GO:0098590 |
query_1 |
GO:CC |
GO:0098590 |
plasma membrane region |
0.0000000 |
194 |
41 |
1408 |
26995 |
41/194 |
1408/26995 |
| GO:0016324 |
query_1 |
GO:CC |
GO:0016324 |
apical plasma membrane |
0.0000000 |
194 |
23 |
461 |
26995 |
23/194 |
461/26995 |
| GO:0016020 |
query_1 |
GO:CC |
GO:0016020 |
membrane |
0.0000000 |
194 |
122 |
10191 |
26995 |
122/194 |
10191/26995 |
| GO:0005886 |
query_1 |
GO:CC |
GO:0005886 |
plasma membrane |
0.0000000 |
194 |
89 |
6367 |
26995 |
89/194 |
6367/26995 |
| GO:0045177 |
query_1 |
GO:CC |
GO:0045177 |
apical part of cell |
0.0000000 |
194 |
23 |
562 |
26995 |
23/194 |
562/26995 |
| GO:0009925 |
query_1 |
GO:CC |
GO:0009925 |
basal plasma membrane |
0.0000029 |
194 |
15 |
312 |
26995 |
15/194 |
312/26995 |
| GO:0045178 |
query_1 |
GO:CC |
GO:0045178 |
basal part of cell |
0.0000064 |
194 |
15 |
331 |
26995 |
15/194 |
331/26995 |
| GO:0016323 |
query_1 |
GO:CC |
GO:0016323 |
basolateral plasma membrane |
0.0000355 |
194 |
13 |
275 |
26995 |
13/194 |
275/26995 |
| GO:0030054 |
query_1 |
GO:CC |
GO:0030054 |
cell junction |
0.0000762 |
194 |
39 |
2260 |
26995 |
39/194 |
2260/26995 |
| GO:0120025 |
query_1 |
GO:CC |
GO:0120025 |
plasma membrane bounded cell projection |
0.0002408 |
194 |
40 |
2460 |
26995 |
40/194 |
2460/26995 |
| GO:0042995 |
query_1 |
GO:CC |
GO:0042995 |
cell projection |
0.0002613 |
194 |
41 |
2564 |
26995 |
41/194 |
2564/26995 |
| GO:0031012 |
query_1 |
GO:CC |
GO:0031012 |
extracellular matrix |
0.0011717 |
194 |
15 |
499 |
26995 |
15/194 |
499/26995 |
| GO:0030312 |
query_1 |
GO:CC |
GO:0030312 |
external encapsulating structure |
0.0012297 |
194 |
15 |
501 |
26995 |
15/194 |
501/26995 |
| GO:0043005 |
query_1 |
GO:CC |
GO:0043005 |
neuron projection |
0.0015662 |
194 |
28 |
1525 |
26995 |
28/194 |
1525/26995 |
| GO:0036477 |
query_1 |
GO:CC |
GO:0036477 |
somatodendritic compartment |
0.0015798 |
194 |
23 |
1103 |
26995 |
23/194 |
1103/26995 |
| GO:0070161 |
query_1 |
GO:CC |
GO:0070161 |
anchoring junction |
0.0019243 |
194 |
18 |
729 |
26995 |
18/194 |
729/26995 |
| GO:0005576 |
query_1 |
GO:CC |
GO:0005576 |
extracellular region |
0.0038707 |
194 |
38 |
2554 |
26995 |
38/194 |
2554/26995 |
| GO:0009986 |
query_1 |
GO:CC |
GO:0009986 |
cell surface |
0.0064805 |
194 |
21 |
1037 |
26995 |
21/194 |
1037/26995 |
| GO:0110165 |
query_1 |
GO:CC |
GO:0110165 |
cellular anatomical entity |
0.0079523 |
194 |
182 |
22653 |
26995 |
182/194 |
22653/26995 |
| GO:0016327 |
query_1 |
GO:CC |
GO:0016327 |
apicolateral plasma membrane |
0.0129450 |
194 |
4 |
27 |
26995 |
4/194 |
27/26995 |
| GO:0045202 |
query_1 |
GO:CC |
GO:0045202 |
synapse |
0.0161906 |
194 |
27 |
1641 |
26995 |
27/194 |
1641/26995 |
| GO:0030425 |
query_1 |
GO:CC |
GO:0030425 |
dendrite |
0.0178150 |
194 |
17 |
783 |
26995 |
17/194 |
783/26995 |
| GO:0097447 |
query_1 |
GO:CC |
GO:0097447 |
dendritic tree |
0.0183813 |
194 |
17 |
785 |
26995 |
17/194 |
785/26995 |
| GO:0030424 |
query_1 |
GO:CC |
GO:0030424 |
axon |
0.0235164 |
194 |
17 |
801 |
26995 |
17/194 |
801/26995 |
| GO:0015629 |
query_1 |
GO:CC |
GO:0015629 |
actin cytoskeleton |
0.0250212 |
194 |
13 |
503 |
26995 |
13/194 |
503/26995 |
| GO:0062023 |
query_1 |
GO:CC |
GO:0062023 |
collagen-containing extracellular matrix |
0.0271177 |
194 |
11 |
371 |
26995 |
11/194 |
371/26995 |
| GO:0043025 |
query_1 |
GO:CC |
GO:0043025 |
neuronal cell body |
0.0325979 |
194 |
16 |
743 |
26995 |
16/194 |
743/26995 |
| GO:0005215 |
query_1 |
GO:MF |
GO:0005215 |
transporter activity |
0.0000000 |
190 |
35 |
1189 |
25063 |
35/190 |
1189/25063 |
| GO:0008514 |
query_1 |
GO:MF |
GO:0008514 |
organic anion transmembrane transporter activity |
0.0000000 |
190 |
17 |
264 |
25063 |
17/190 |
264/25063 |
| GO:0022857 |
query_1 |
GO:MF |
GO:0022857 |
transmembrane transporter activity |
0.0000001 |
190 |
31 |
1087 |
25063 |
31/190 |
1087/25063 |
| GO:0022804 |
query_1 |
GO:MF |
GO:0022804 |
active transmembrane transporter activity |
0.0000007 |
190 |
19 |
435 |
25063 |
19/190 |
435/25063 |
| GO:0046943 |
query_1 |
GO:MF |
GO:0046943 |
carboxylic acid transmembrane transporter activity |
0.0000017 |
190 |
13 |
189 |
25063 |
13/190 |
189/25063 |
| GO:0005342 |
query_1 |
GO:MF |
GO:0005342 |
organic acid transmembrane transporter activity |
0.0000018 |
190 |
13 |
190 |
25063 |
13/190 |
190/25063 |
| GO:0005319 |
query_1 |
GO:MF |
GO:0005319 |
lipid transporter activity |
0.0000096 |
190 |
12 |
179 |
25063 |
12/190 |
179/25063 |
| GO:0015291 |
query_1 |
GO:MF |
GO:0015291 |
secondary active transmembrane transporter activity |
0.0000377 |
190 |
14 |
290 |
25063 |
14/190 |
290/25063 |
| GO:0005515 |
query_1 |
GO:MF |
GO:0005515 |
protein binding |
0.0000424 |
190 |
116 |
10460 |
25063 |
116/190 |
10460/25063 |
| GO:0042910 |
query_1 |
GO:MF |
GO:0042910 |
xenobiotic transmembrane transporter activity |
0.0002910 |
190 |
6 |
38 |
25063 |
6/190 |
38/25063 |
| GO:0015173 |
query_1 |
GO:MF |
GO:0015173 |
aromatic amino acid transmembrane transporter activity |
0.0004768 |
190 |
4 |
10 |
25063 |
4/190 |
10/25063 |
| GO:0008028 |
query_1 |
GO:MF |
GO:0008028 |
monocarboxylic acid transmembrane transporter activity |
0.0006871 |
190 |
7 |
69 |
25063 |
7/190 |
69/25063 |
| GO:0005290 |
query_1 |
GO:MF |
GO:0005290 |
L-histidine transmembrane transporter activity |
0.0012543 |
190 |
3 |
4 |
25063 |
3/190 |
4/25063 |
| GO:0140333 |
query_1 |
GO:MF |
GO:0140333 |
glycerophospholipid flippase activity |
0.0022194 |
190 |
4 |
14 |
25063 |
4/190 |
14/25063 |
| GO:0140327 |
query_1 |
GO:MF |
GO:0140327 |
flippase activity |
0.0051837 |
190 |
4 |
17 |
25063 |
4/190 |
17/25063 |
| GO:0001540 |
query_1 |
GO:MF |
GO:0001540 |
amyloid-beta binding |
0.0080306 |
190 |
6 |
66 |
25063 |
6/190 |
66/25063 |
| GO:0050839 |
query_1 |
GO:MF |
GO:0050839 |
cell adhesion molecule binding |
0.0112496 |
190 |
11 |
292 |
25063 |
11/190 |
292/25063 |
| GO:0015175 |
query_1 |
GO:MF |
GO:0015175 |
neutral L-amino acid transmembrane transporter activity |
0.0118318 |
190 |
5 |
42 |
25063 |
5/190 |
42/25063 |
| GO:0015318 |
query_1 |
GO:MF |
GO:0015318 |
inorganic molecular entity transmembrane transporter activity |
0.0323750 |
190 |
16 |
657 |
25063 |
16/190 |
657/25063 |
| GO:1901618 |
query_1 |
GO:MF |
GO:1901618 |
organic hydroxy compound transmembrane transporter activity |
0.0373100 |
190 |
5 |
53 |
25063 |
5/190 |
53/25063 |
| GO:0140326 |
query_1 |
GO:MF |
GO:0140326 |
ATPase-coupled intramembrane lipid transporter activity |
0.0417784 |
190 |
4 |
28 |
25063 |
4/190 |
28/25063 |
| GO:0140830 |
query_1 |
GO:MF |
GO:0140830 |
L-glutamine, sodium:proton antiporter activity |
0.0420366 |
190 |
2 |
2 |
25063 |
2/190 |
2/25063 |
| GO:0017147 |
query_1 |
GO:MF |
GO:0017147 |
Wnt-protein binding |
0.0481766 |
190 |
4 |
29 |
25063 |
4/190 |
29/25063 |
| KEGG:05200 |
query_1 |
KEGG |
KEGG:05200 |
Pathways in cancer |
0.0348500 |
86 |
14 |
543 |
9330 |
14/86 |
543/9330 |
| KEGG:05226 |
query_1 |
KEGG |
KEGG:05226 |
Gastric cancer |
0.0387416 |
86 |
7 |
150 |
9330 |
7/86 |
150/9330 |
| REAC:R-MMU-382551 |
query_1 |
REAC |
REAC:R-MMU-382551 |
Transport of small molecules |
0.0000003 |
84 |
25 |
628 |
8405 |
25/84 |
628/8405 |
| REAC:R-MMU-425407 |
query_1 |
REAC |
REAC:R-MMU-425407 |
SLC-mediated transmembrane transport |
0.0000005 |
84 |
15 |
207 |
8405 |
15/84 |
207/8405 |
| REAC:R-MMU-352230 |
query_1 |
REAC |
REAC:R-MMU-352230 |
Amino acid transport across the plasma membrane |
0.0027347 |
84 |
5 |
29 |
8405 |
5/84 |
29/8405 |
| REAC:R-MMU-425393 |
query_1 |
REAC |
REAC:R-MMU-425393 |
Transport of inorganic cations/anions and amino acids/oligopeptides |
0.0115187 |
84 |
7 |
93 |
8405 |
7/84 |
93/8405 |
| REAC:R-MMU-556833 |
query_1 |
REAC |
REAC:R-MMU-556833 |
Metabolism of lipids |
0.0430149 |
84 |
16 |
570 |
8405 |
16/84 |
570/8405 |
| TF:M07289 |
query_1 |
TF |
TF:M07289 |
Factor: GKLF; motif: NNNRGGNGNGGSN |
0.0000028 |
191 |
169 |
15234 |
21629 |
169/191 |
15234/21629 |
| TF:M10375 |
query_1 |
TF |
TF:M10375 |
Factor: Sp1; motif: GGNGGGGGNGGGGGMGGGGCNGGG |
0.0000176 |
191 |
133 |
10720 |
21629 |
133/191 |
10720/21629 |
| TF:M10276_1 |
query_1 |
TF |
TF:M10276_1 |
Factor: Kaiso; motif: SARNYCTCGCGAGAN; match class: 1 |
0.0000706 |
191 |
125 |
9977 |
21629 |
125/191 |
9977/21629 |
| TF:M02933 |
query_1 |
TF |
TF:M02933 |
Factor: ZF5; motif: GYCGCGCARNGCNN |
0.0001358 |
191 |
120 |
9499 |
21629 |
120/191 |
9499/21629 |
| TF:M02023 |
query_1 |
TF |
TF:M02023 |
Factor: MAZ; motif: NKGGGAGGGGRGGR |
0.0002606 |
191 |
105 |
7941 |
21629 |
105/191 |
7941/21629 |
| TF:M02933_1 |
query_1 |
TF |
TF:M02933_1 |
Factor: ZF5; motif: GYCGCGCARNGCNN; match class: 1 |
0.0003833 |
191 |
84 |
5832 |
21629 |
84/191 |
5832/21629 |
| TF:M07289_1 |
query_1 |
TF |
TF:M07289_1 |
Factor: GKLF; motif: NNNRGGNGNGGSN; match class: 1 |
0.0007775 |
191 |
130 |
10917 |
21629 |
130/191 |
10917/21629 |
| TF:M00333 |
query_1 |
TF |
TF:M00333 |
Factor: ZF5; motif: NRNGNGCGCGCWN |
0.0013384 |
191 |
157 |
14398 |
21629 |
157/191 |
14398/21629 |
| TF:M00333_1 |
query_1 |
TF |
TF:M00333_1 |
Factor: ZF5; motif: NRNGNGCGCGCWN; match class: 1 |
0.0019685 |
191 |
131 |
11183 |
21629 |
131/191 |
11183/21629 |
| TF:M10209 |
query_1 |
TF |
TF:M10209 |
Factor: E2F-1; motif: GNGGGCGGGRMN |
0.0022123 |
191 |
155 |
14210 |
21629 |
155/191 |
14210/21629 |
| TF:M00716 |
query_1 |
TF |
TF:M00716 |
Factor: ZF5; motif: GSGCGCGR |
0.0035464 |
191 |
159 |
14823 |
21629 |
159/191 |
14823/21629 |
| TF:M07436 |
query_1 |
TF |
TF:M07436 |
Factor: WT1; motif: NNGGGNGGGSGN |
0.0037644 |
191 |
99 |
7691 |
21629 |
99/191 |
7691/21629 |
| TF:M07326_1 |
query_1 |
TF |
TF:M07326_1 |
Factor: Mef-2A; motif: GNNYTAAAWATAR; match class: 1 |
0.0038961 |
191 |
5 |
26 |
21629 |
5/191 |
26/21629 |
| TF:M02023_1 |
query_1 |
TF |
TF:M02023_1 |
Factor: MAZ; motif: NKGGGAGGGGRGGR; match class: 1 |
0.0045374 |
191 |
63 |
4130 |
21629 |
63/191 |
4130/21629 |
| TF:M10209_1 |
query_1 |
TF |
TF:M10209_1 |
Factor: E2F-1; motif: GNGGGCGGGRMN; match class: 1 |
0.0045908 |
191 |
125 |
10616 |
21629 |
125/191 |
10616/21629 |
| TF:M00803 |
query_1 |
TF |
TF:M00803 |
Factor: E2F; motif: GGCGSG |
0.0054322 |
191 |
123 |
10412 |
21629 |
123/191 |
10412/21629 |
| TF:M01118 |
query_1 |
TF |
TF:M01118 |
Factor: WT1; motif: SMCNCCNSC |
0.0055003 |
191 |
102 |
8071 |
21629 |
102/191 |
8071/21629 |
| TF:M00803_1 |
query_1 |
TF |
TF:M00803_1 |
Factor: E2F; motif: GGCGSG; match class: 1 |
0.0062111 |
191 |
92 |
7031 |
21629 |
92/191 |
7031/21629 |
| TF:M07040 |
query_1 |
TF |
TF:M07040 |
Factor: GKLF; motif: NNRRGRRNGNSNNN |
0.0085054 |
191 |
143 |
12890 |
21629 |
143/191 |
12890/21629 |
| TF:M01104 |
query_1 |
TF |
TF:M01104 |
Factor: MOVO-B; motif: GNGGGGG |
0.0102348 |
191 |
97 |
7635 |
21629 |
97/191 |
7635/21629 |
| TF:M10276 |
query_1 |
TF |
TF:M10276 |
Factor: Kaiso; motif: SARNYCTCGCGAGAN |
0.0131366 |
191 |
147 |
13469 |
21629 |
147/191 |
13469/21629 |
| TF:M02742 |
query_1 |
TF |
TF:M02742 |
Factor: E2F-2; motif: NNNANGGCGCGCNNN |
0.0136810 |
191 |
55 |
3530 |
21629 |
55/191 |
3530/21629 |
| TF:M01240 |
query_1 |
TF |
TF:M01240 |
Factor: BEN; motif: CAGCGRNV |
0.0174652 |
191 |
167 |
16194 |
21629 |
167/191 |
16194/21629 |
| TF:M00716_1 |
query_1 |
TF |
TF:M00716_1 |
Factor: ZF5; motif: GSGCGCGR; match class: 1 |
0.0185489 |
191 |
135 |
12044 |
21629 |
135/191 |
12044/21629 |
| TF:M10375_1 |
query_1 |
TF |
TF:M10375_1 |
Factor: Sp1; motif: GGNGGGGGNGGGGGMGGGGCNGGG; match class: 1 |
0.0280992 |
191 |
93 |
7376 |
21629 |
93/191 |
7376/21629 |
| TF:M00665 |
query_1 |
TF |
TF:M00665 |
Factor: Sp3; motif: ASMCTTGGGSRGGG |
0.0389990 |
191 |
91 |
7220 |
21629 |
91/191 |
7220/21629 |
| TF:M02089 |
query_1 |
TF |
TF:M02089 |
Factor: E2F-3; motif: GGCGGGN |
0.0477466 |
191 |
127 |
11263 |
21629 |
127/191 |
11263/21629 |
| WP:WP4344 |
query_1 |
WP |
WP:WP4344 |
Sphingolipid metabolism overview |
0.0060915 |
52 |
4 |
24 |
4496 |
4/52 |
24/4496 |
| WP:WP5298 |
query_1 |
WP |
WP:WP5298 |
Dravet syndrome Scn1a A1783V point mutation model |
0.0066423 |
52 |
6 |
72 |
4496 |
6/52 |
72/4496 |
| WP:WP4690 |
query_1 |
WP |
WP:WP4690 |
Sphingolipid metabolism integrated pathway |
0.0071900 |
52 |
4 |
25 |
4496 |
4/52 |
25/4496 |